Evercode TCR Mega
/in Evercode, Parse, Partners/by Harshita SharmaPRODUCTS
Evercode TCR Mega
1M CELLS
96 SAMPLES
Unlock scalable immune profiling capabilities with Evercode™ TCR Mega, empowering you to analyze up to 1 million cells at a time. In the intricate landscape of immunity, comprehensive data is key. With Evercode™ TCR Mega, you can achieve unparalleled resolution in studying TCR clonotype diversity and T-cell states.
Benefit from the expansive scale of Evercode™ TCR Mega, which offers a host of advantages alongside the wealth of information provided by WT profiles. Whether you’re exploring immune responses, investigating cellular dynamics, or delving into immunotherapy research, Evercode™ TCR Mega equips you with the tools to navigate the complexities of the immune system with confidence and precision.
EXPONENTIALLY
SCALABLE
Evercode's combinatorial barcoding
enables you to dramatically
scale up the cells and samples per experiment.
NO INSTRUMENT
REQUIRED
If you have a centrifuge,
thermal cycler,
and some pipettes,
you’re ready to go.
UNMATCHED DATA QUALITY
Better detect lowly expressed
genes and avoid ambient RNA common
in droplet-based
single cell sequencing.
WORKS WITH FIXED CELLS AND NUCLEI
Fix and store samples as they come in for up to 6 months and then run together later on your schedule. Ideal for cross-site collaborations.
Experience the power of scale with Evercode. Conduct experiments with up to 1 million cells in a single run, unlocking the immense diversity within the immune repertoire like never before. With Evercode, identify an extensive array of TCR clonotypes, providing unparalleled insights into immune function and response.
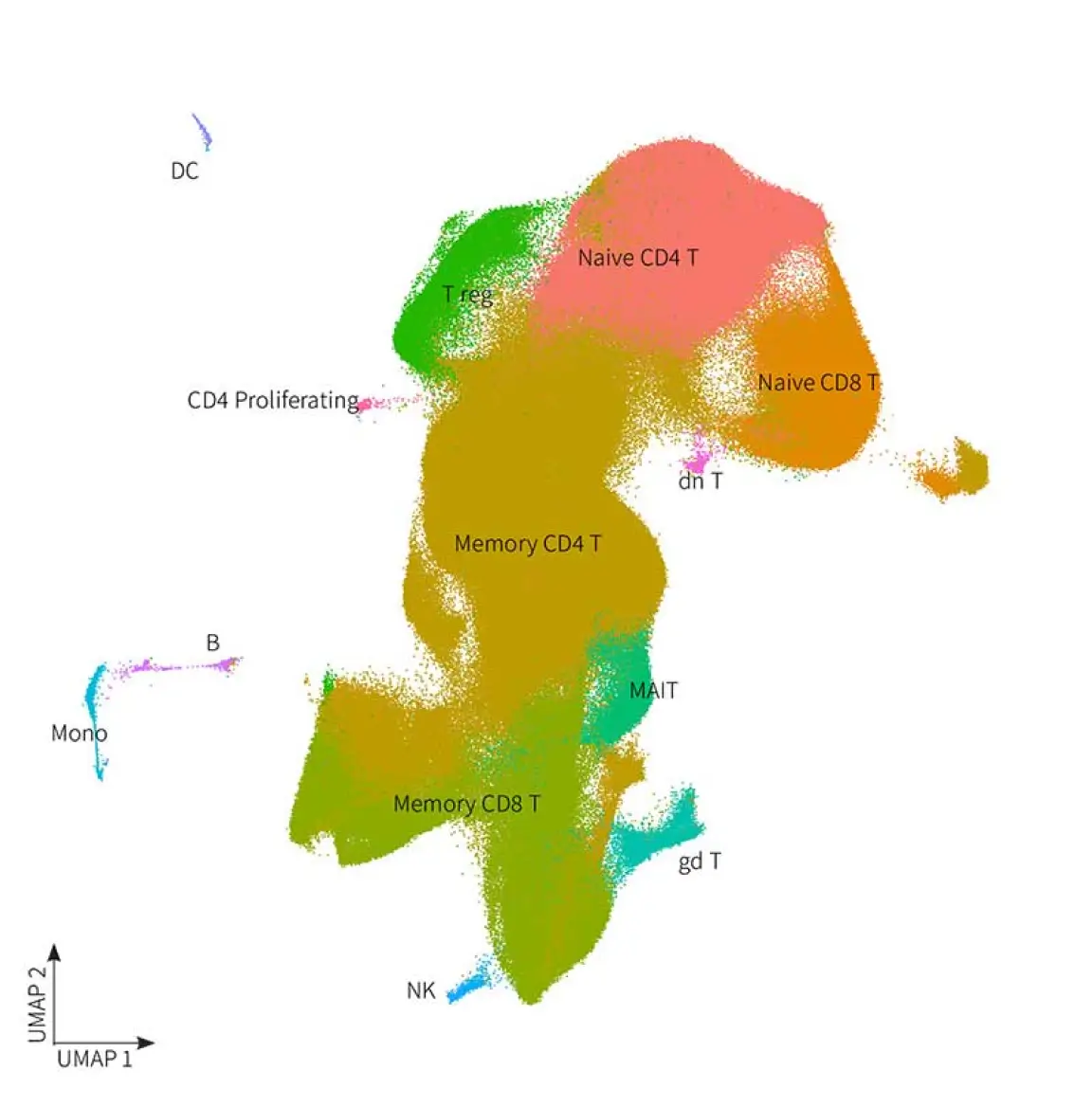
Not only can Evercode detect clonotypes, but it also offers the capability to map them to T cell subtype clusters. By analyzing gene expression profiles from nearly 1 million T cells isolated from PBMCs, Evercode enables the classification of cells into distinct T cell subtypes. This comprehensive approach ensures a deeper understanding of immune dynamics and facilitates more nuanced analysis of immune responses.
Experience outstanding TCR detection in both activated and primary T cells with our cutting-edge profiling techniques. In our investigation, T cells derived from peripheral blood mononuclear cells (PBMCs) underwent two distinct conditions: direct profiling (Primary) and activation through culture for 3 days with CD3/CD28 beads and IL-2 (Activated).
Our results demonstrate robust TCR detection in both experimental conditions, highlighting the versatility and reliability of our methodology across diverse T cell states. Whether profiling primary T cells or their activated counterparts, our approach ensures accurate and comprehensive characterization of the TCR repertoire, providing invaluable insights into immune responses and cellular dynamics.
Trust in our advanced profiling techniques to deliver precise and high-fidelity data, enabling a deeper understanding of T cell biology and immunological processes. With our innovative methodologies, delve into the complexities of the immune system with confidence and clarity, paving the way for transformative discoveries in immunology.
Immerse yourself in unparalleled detection of the immune repertoire with the Evercode™ TCR kit, meticulously crafted to offer the most comprehensive profile attainable. In a groundbreaking study spanning 8 donors, our kit revealed the identification of nearly 500,000 unique beta chain clonotypes.
Notably, the majority of these clonotypes were deemed rare, emphasizing the remarkable sensitivity and depth of our detection capabilities. With the Evercode™ TCR kit at your disposal, you gain the ability to unveil the complete spectrum of immune diversity, from commonplace to exceptionally rare clonotypes. This empowers you to delve into the intricacies of immune responses with unparalleled precision and breadth, opening new avenues for exploration and discovery in immunology.

Let’s Find You an Application That Helps Your Research
Get a call from your local Decode Science representative to help you find the best fit genomics products for you.
Or give us a call at: