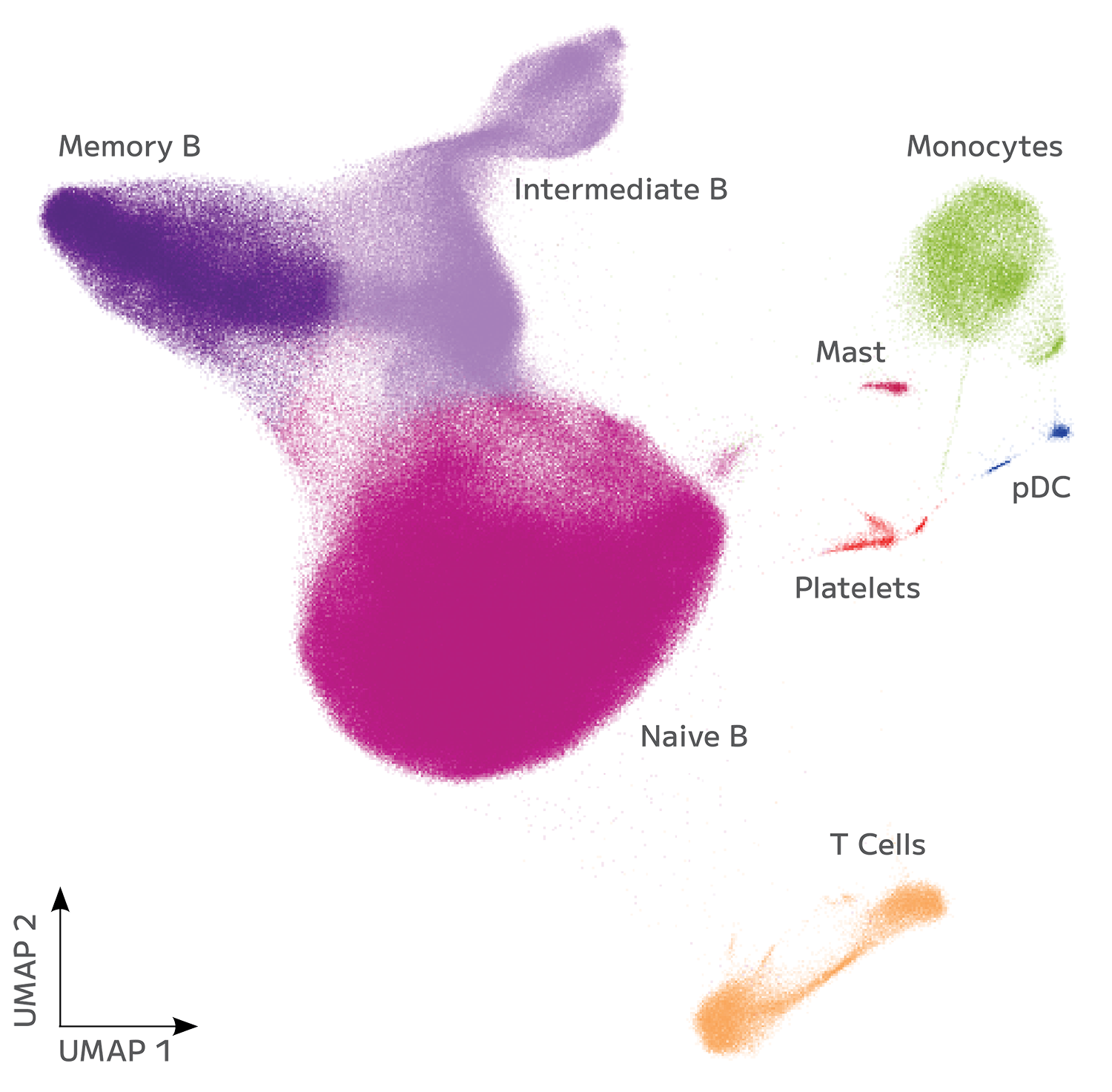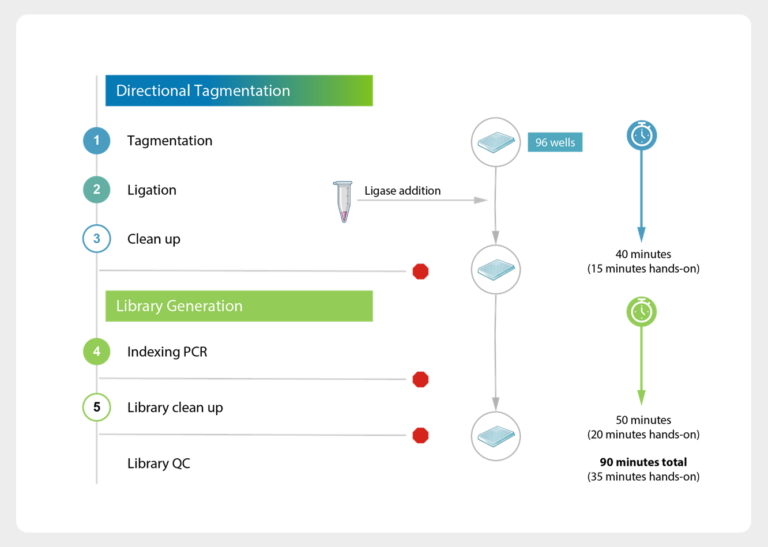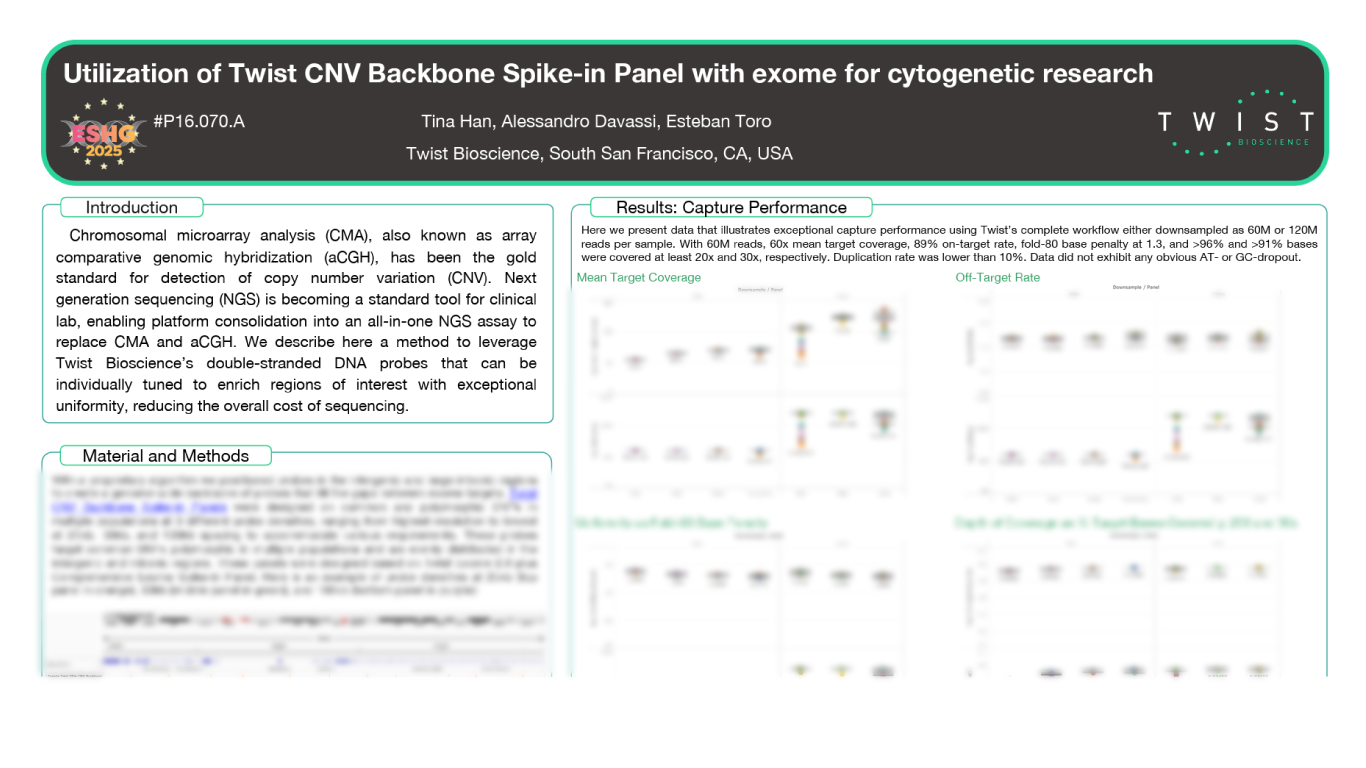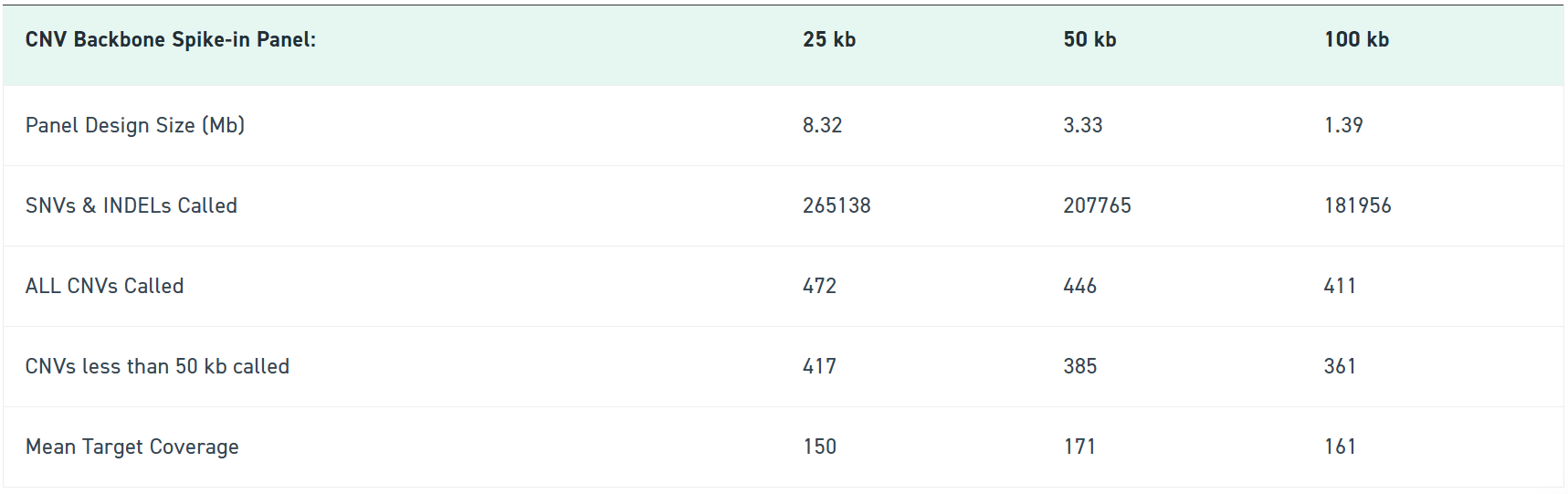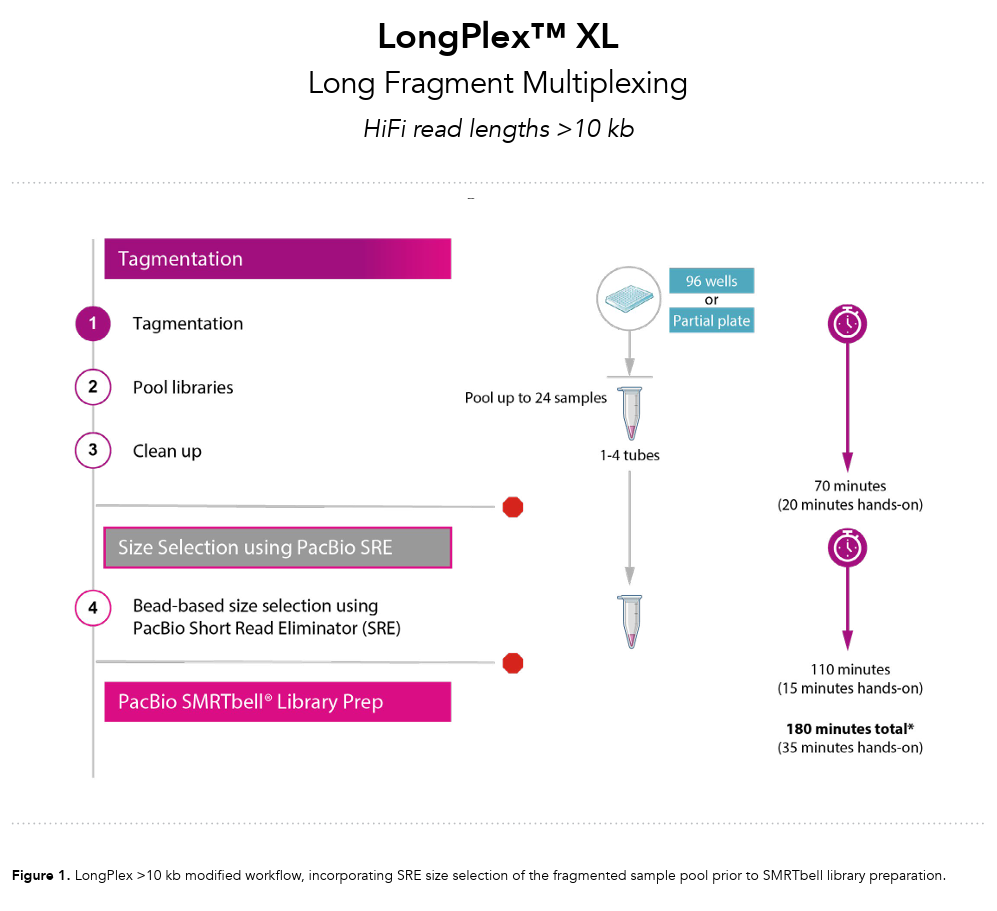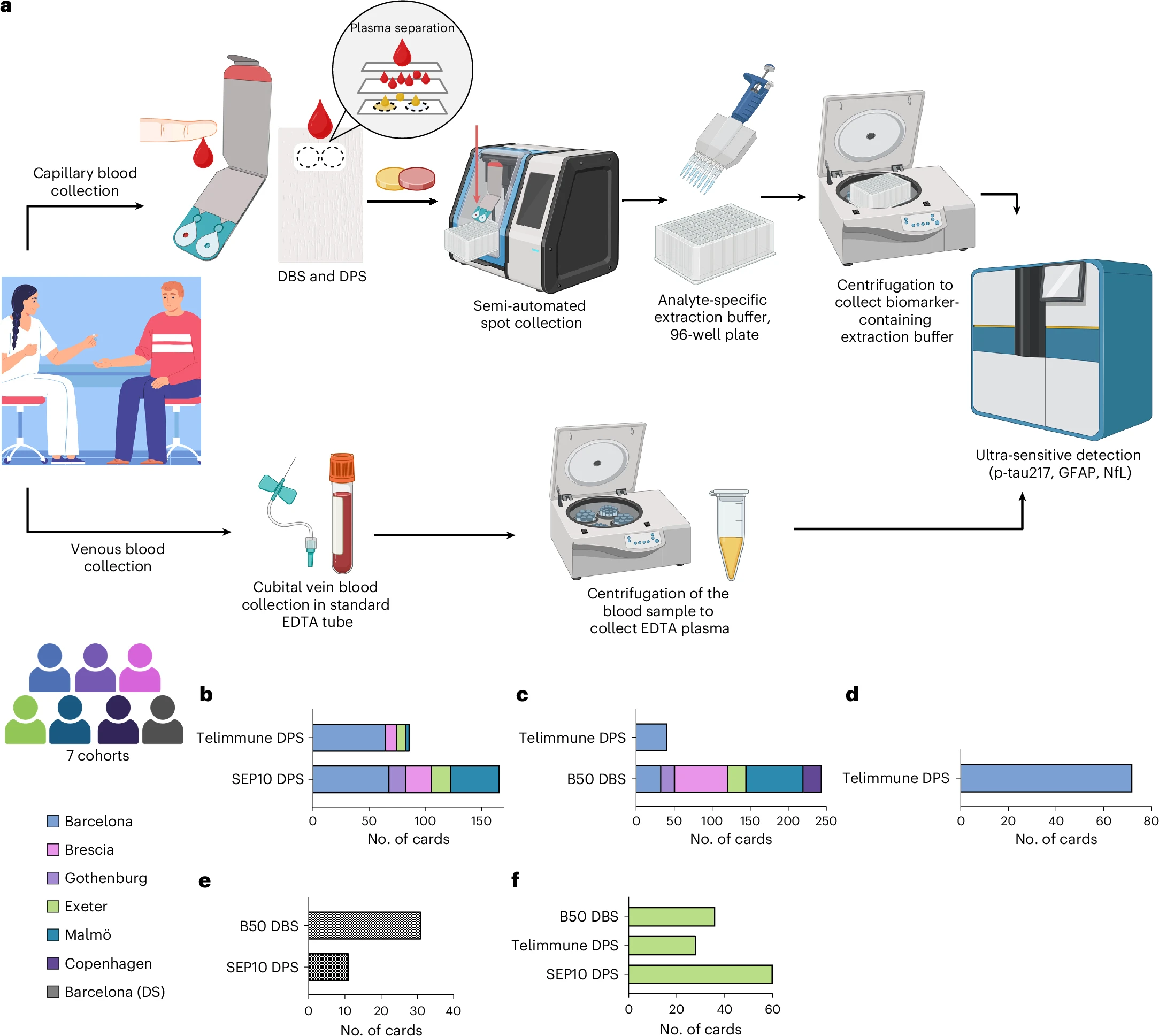
Huber, H., Montoliu-Gaya, L., Brum, W.S. et al. A minimally invasive dried blood spot biomarker test for the detection of Alzheimer’s disease pathology. Nat Med (2026). https://doi.org/10.1038/s41591-025-04080-0
The Simoa Dry Blood Extraction Kit is a validated, device-agnostic solution for extracting analytes from dried plasma and dried blood spot (DPS/DBS) samples for use with Simoa® assays. It enables consistent, reproducible recovery of low-abundance biomarkers while maintaining the femtogram-level sensitivity required for clinical and translational research.
Designed to support decentralized and remote sample collection, the kit simplifies pre-analytical workflows without compromising data quality. It is well suited for longitudinal studies, multi-site trials, and settings where traditional venous sampling and cold-chain logistics are impractical.