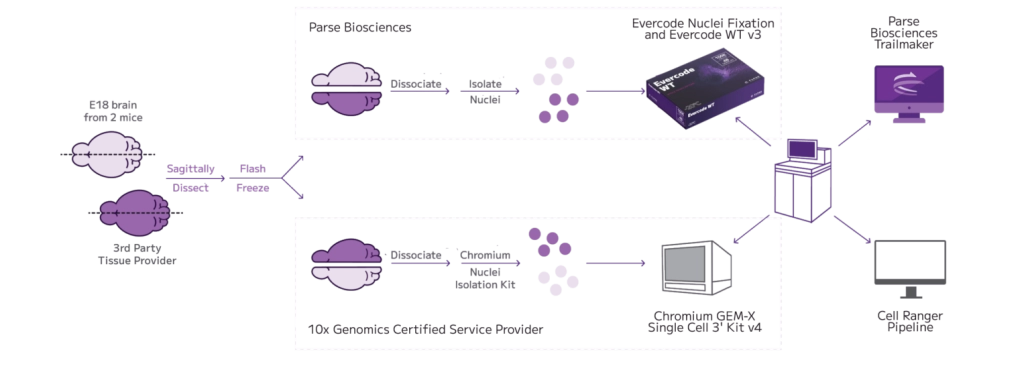
Stronger CNV Detection — Without Changing Your Exome Workflow.
Large copy number variations can be easy to miss when exome sequencing focuses on exonic regions – leading to blind spots, repeat testing, and extra time interpreting unclear data. By spiking in Twist’s CNV Backbone Panels, you give your exome run the evenly spaced genomic coverage it needs to reliably surface clinically relevant CNVs — especially the ones standard exomes struggle with.
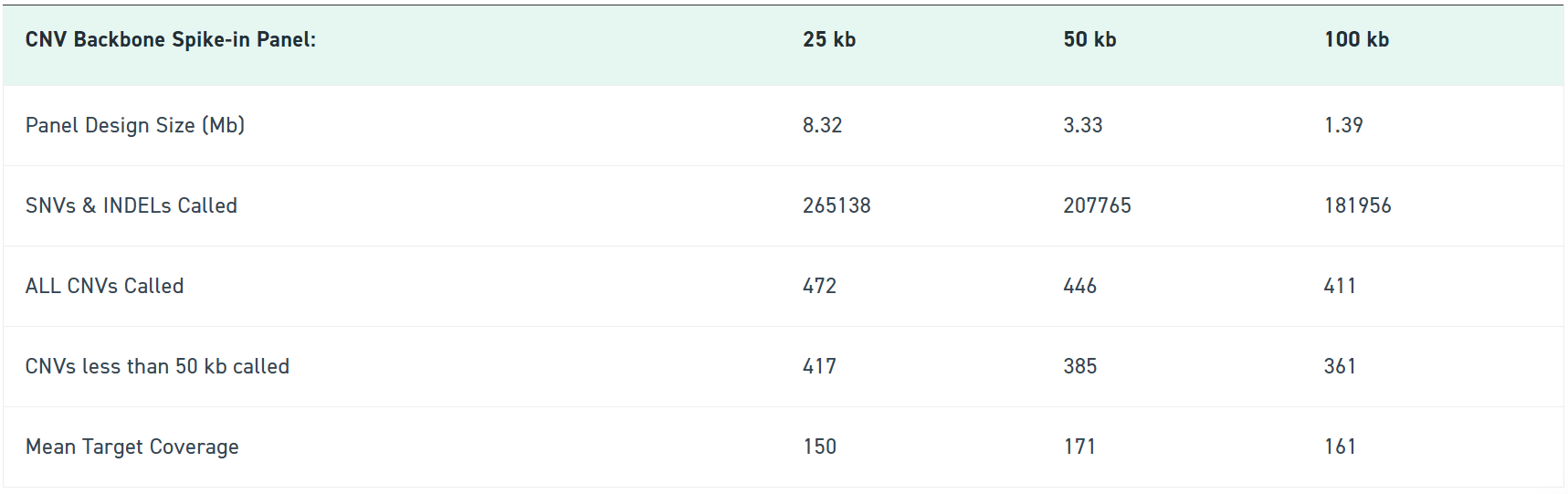
Available in 100 kb, 50 kb, and 25 kb resolutions, the CNV Backbone Panels strengthen your detection sensitivity while keeping your workflow identical. Blend it into your existing exome panel, follow your standard Twist enrichment protocol, and immediately get more confident CNV calls backed by consistent probe tiling across intergenic regions.