TCR Library
/in Partners, Synbio Libraries, Twist/by Harshita SharmaAdoptive cell therapy (ACT) has transformed treatment options for patients with advanced or refractory cancers by redirecting the immune system to recognize and eliminate malignant cells.
Engineered TCR therapies represent a major branch of ACT, using synthetic or optimized T-cell receptors to target tumor-specific antigens. The workflow typically begins with sequencing the tumor biopsy to identify actionable mutations and the peripheral blood to map the patient’s TCR repertoire.
TCR repertoire profiling can be performed using bulk or single-cell sequencing, each offering distinct advantages:
Bulk sequencing provides broad coverage of the repertoire and captures large numbers of clonotypes, but does not preserve α–β chain pairing.
Single-cell sequencing preserves α–β receptor pairing and full receptor architecture, but at a much lower throughput than bulk methods.
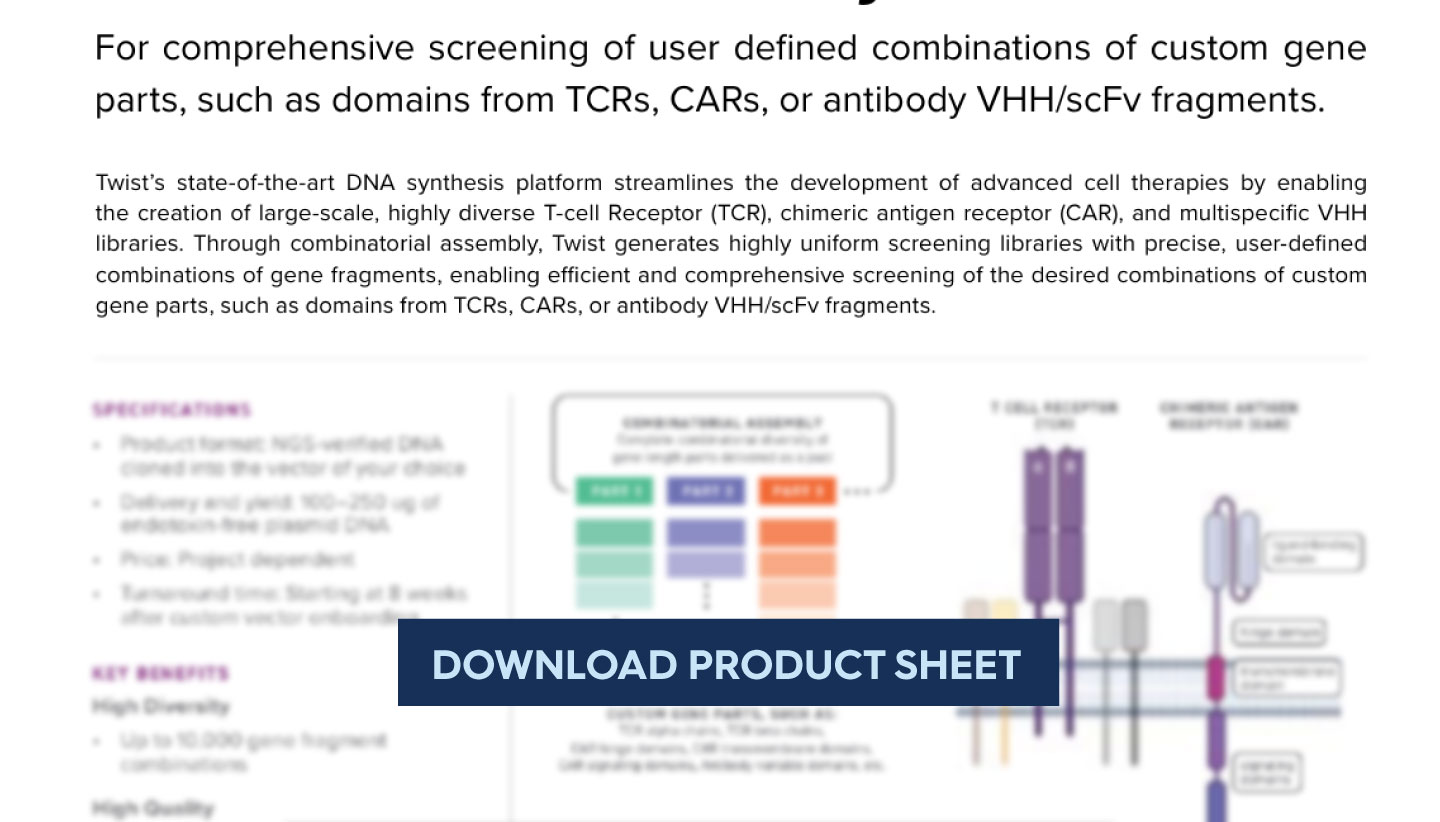
Twist’s combinatorial TCR library platform gives you the power to systematically reshape and expand your TCR landscape. Instead of being limited to naturally occurring α–β pairs, you can mix and match chains to uncover novel receptor combinations with therapeutic potential.
This approach pairs especially well with bulk repertoire sequencing, where α–β pairing is not preserved. Combinatorial assembly fills that gap by generating all possible pairings across defined alpha and beta chains.
Example:
A library built from 4 α-chains and 4 β-chains yields 16 unique α–β combinations — complete coverage, not a partial subset.
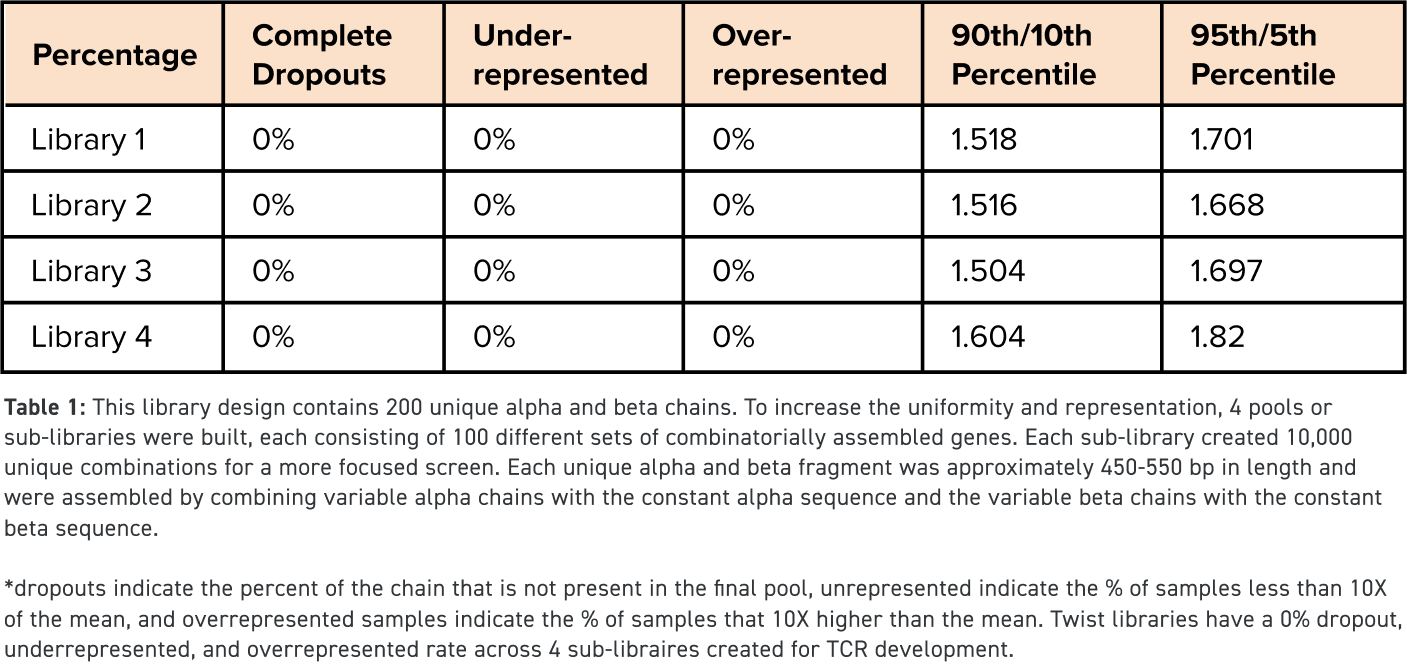
High Diversity & High Quality
Up to 10,000 combinatorial gene fragment combinations per library
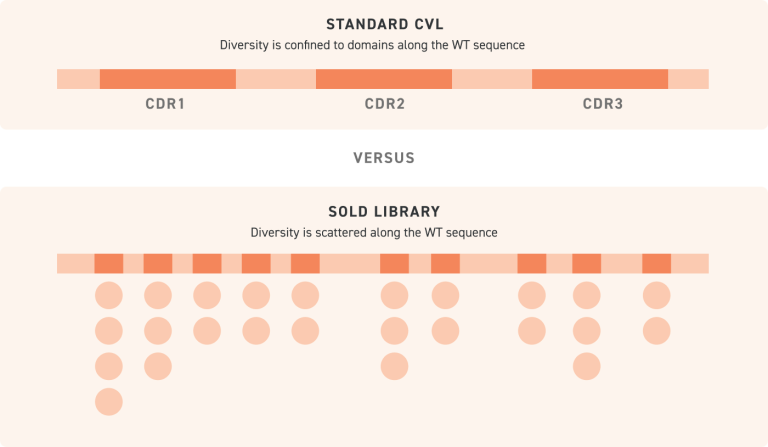
NGS-verified with >90% of expected variants represented within 10× of the mean
Design Exactly What You Need
Define your own α–β chain combinations for fully customized TCR libraries
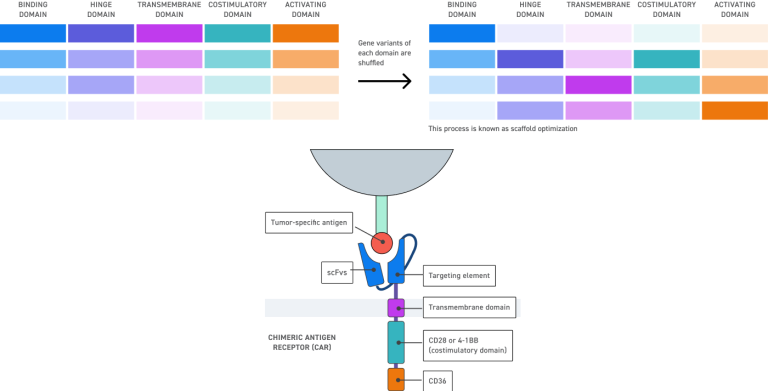
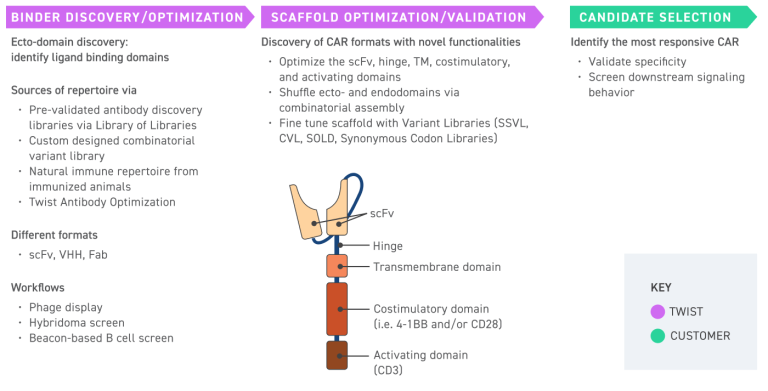
CAR designs — configurable hinge, transmembrane, and signaling domains
Flexible, Scalable Architecture
Supports inserts up to 1.5 kb
Enables library-scale diversity across multiple sequence elements, tailored to your throughput and screening needs
Unlock with quick sign up!
TCR discovery isn’t limited by ideas — it’s limited by how fast you can screen and rebuild candidates. The real bottleneck comes from needing to evaluate large numbers of alpha–beta pairings to find the rare binders that actually matter.
Combinatorial TCR Libraries break that bottleneck.
By shuffling alpha and beta chains across all possible pairings, these libraries recreate a dramatically broader search space than what exists in the native repertoire. Instead of being restricted to the pairings captured in your sequencing data, you can now generate thousands of rationally designed combinations in a single build.
This enables teams to:
Scale screening throughput by producing many more testable TCR variants
Expand functional diversity beyond naturally occurring pairings
Capture rare or non-intuitive binders that would never appear in arrayed or bulk sequencing formats
Shorten discovery timelines by moving directly from variant design → library synthesis → high-throughput screening

Chris Wicky
Clinical Genomics Manager - ANZ
& Country Manager - NZ
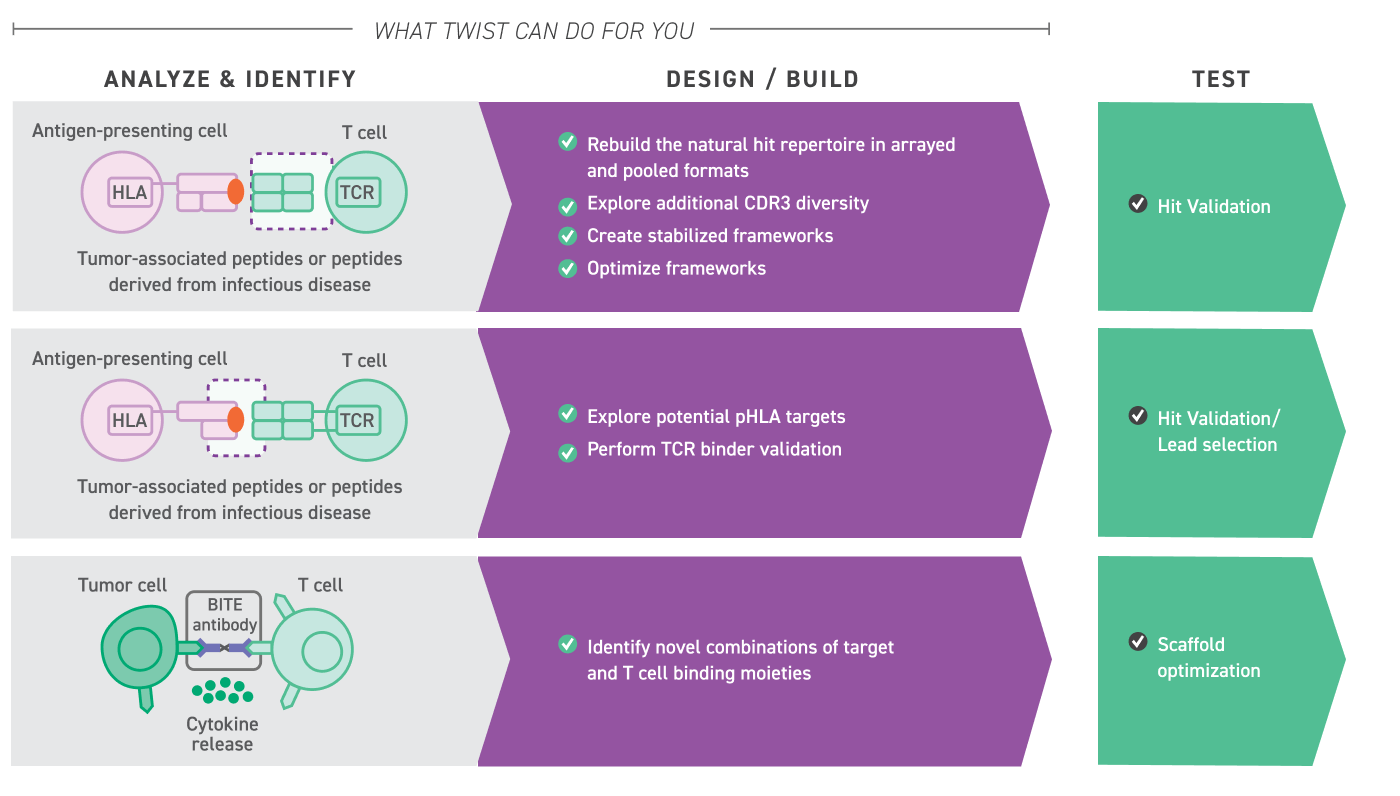
Partner with Twist Bioscience to accelerate the identification and development of advanced cell therapies using large-scale, highly diverse T-cell receptor (TCR) libraries.
Twist provides highly uniform, NGS-verified libraries composed of precise, user-defined combinations of alpha and beta gene fragments. These libraries enable:
Comprehensive Screening: Efficiently test all desired TCR combinations to uncover high-affinity, functional receptors.
Customizable Design: Define specific alpha-beta pairings to target particular antigens or therapeutic applications.
High Uniformity: Libraries are optimized for reproducibility and coverage, ensuring consistent performance across screens.
This approach streamlines TCR discovery, empowering researchers to explore broader sequence diversity and accelerate the development of next-generation engineered TCR therapies.

NGS
Raise confidence in variant detection with superior target enrichment solutions

Oligo Pools
Precision, uniformity, and flexibility for results you can trust

Synthetic Controls
Synthetic RNA and DNA standards for assay development

Libraries
Identify more hits and streamline screening with Twist's precise Variant Libraries