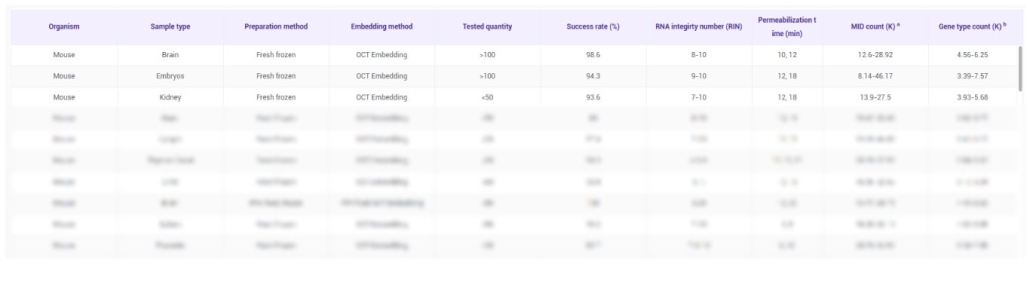
Our STOmics validated tissue list provides researchers with a comprehensive reference of hundreds of tissue types successfully tested using Stereo-seq, the cutting-edge spatial transcriptomics technology. Each tissue entry includes detailed sample information, experimental parameters, and test results, allowing scientists to make informed decisions before starting their single-cell or spatial transcriptomics experiments. By consulting this list, you can ensure compatibility with your tissue samples and streamline your experimental design.
The list not only highlights tissue types that have been validated but also provides insights into the experimental conditions that yielded the most reliable results. Researchers can leverage this information to optimize sample preparation, sequencing protocols, and data quality control measures. This reduces trial-and-error, saves valuable time and resources, and ensures reproducibility across studies. It is an essential tool for anyone planning to use Stereo-seq for spatial gene expression profiling.
In addition, our STOmics validated tissue list supports better planning for large-scale studies and comparative analyses. By providing a centralized reference for tissue performance, it helps guide tissue selection, anticipate potential challenges, and maximize experimental success. Whether you are exploring new tissue types or scaling up existing workflows, this validated tissue list is your key resource for robust, high-quality spatial transcriptomics research.
Notices:
The STOmics validated tissue list was generated using standard tissue and sample types, all of which are frozen. Each tissue sample had an area of less than 1 cm² and was sectioned at a thickness of 10 μm. Most experiments were performed using the Stereo-seq Transcriptomics Kit V1.2, with a few using V1.1. Sequencing depth ranged from 1–3 G reads per sample, and data were processed using the Stereo-seq Analysis Workflow (SAW) versions V2.1.0–V5.1.3. Testing was conducted between 2020 and 2022.
Please note that all test parameters and results are highly dependent on the tissue and sample type. This information should be used as a reference guide to help design and optimize your own experiments, rather than as definitive outcomes for all samples.
Key parameters included in the list:
MID (K): Bin200_Median_MID in thousands
GENETYPE (K): Bin200_Median_Genetype in thousands

ANZ Market Manager - Research Genomics
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.