Key Takeaways
Analyze 10 million cells across 1,152 samples in a single experiment
Increase statistical power by profiling more cells per sample
Capture detailed cellular responses to perturbations and drug treatments
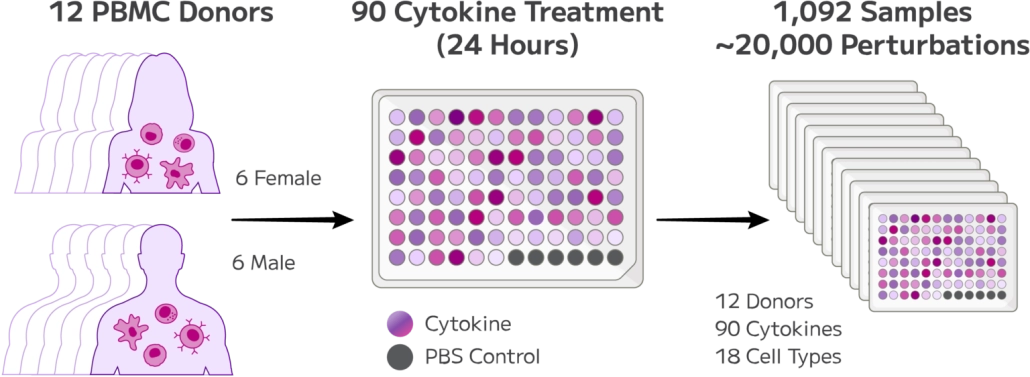
Figure 1: Experimental Design Overview
Approximately 10 million PBMCs from 12 healthy donors were treated with 90 different cytokines in a single GigaLab experiment, covering 1,092 experimental conditions.
Cells were thawed, washed, and seeded at 1 million cells per well across 12 plates. After 24-hour cytokine treatment, cells were fixed, barcoded, and processed for whole transcriptome sequencing. Libraries were sequenced on the Ultima Genomics platform, achieving ~31,000 reads per cell, with 62.45% cell retention after barcoding.
After data processing with the Parse Analysis Pipeline v1.4.0, integration, and classification, 9,697,974 cells across 18 immune cell types were identified—including rare populations that are typically missed in smaller experiments. Each condition yielded a median of 7,400 cells, enabling high-resolution analysis of immune responses.
Differential expression analysis identified how cytokines influenced gene activity across cell types. Many cytokines triggered strong transcriptional responses, with over 50 genes upregulated per treatment.
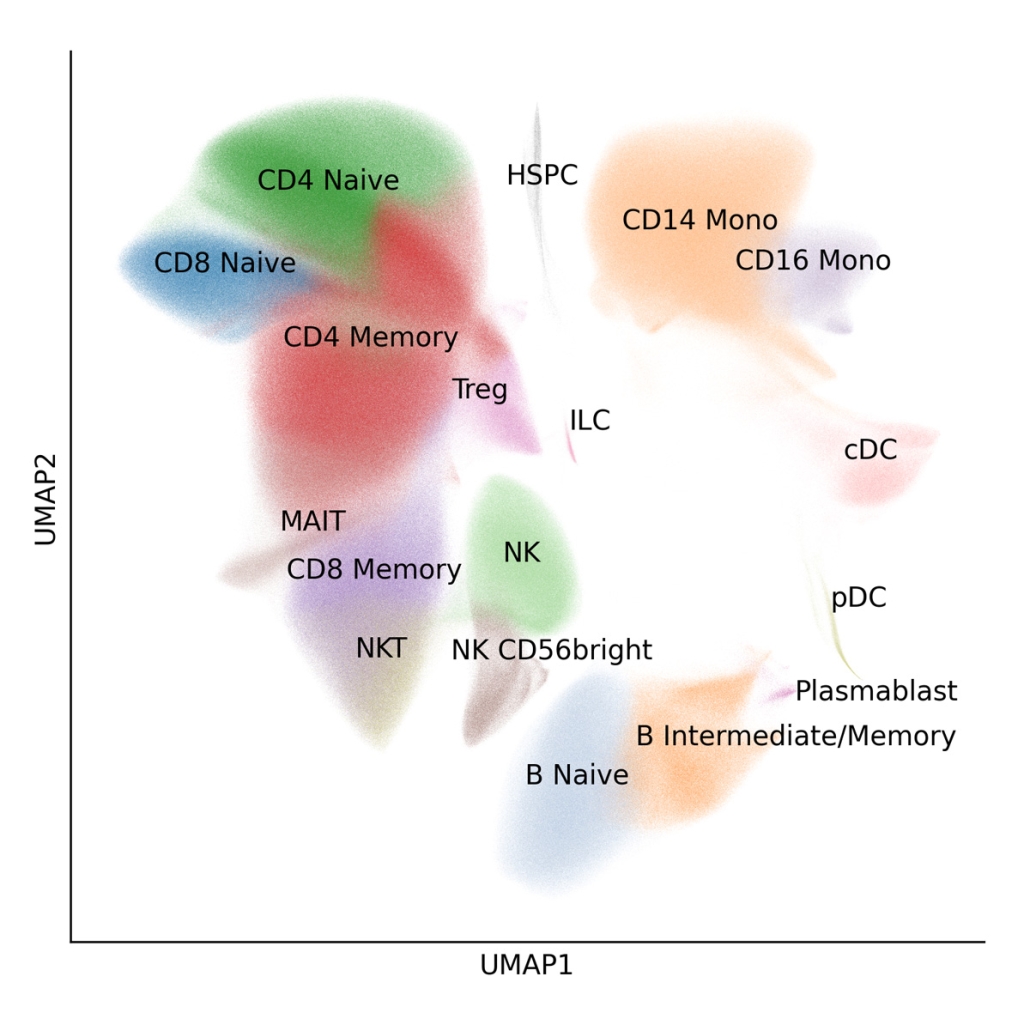
Figure 2: Single-Cell UMAP Overview
9,697,974 PBMCs from 12 donors were integrated with Harmony, clustered using Scanpy, and manually annotated, revealing 18 immune cell types present across all donors and experimental conditions.
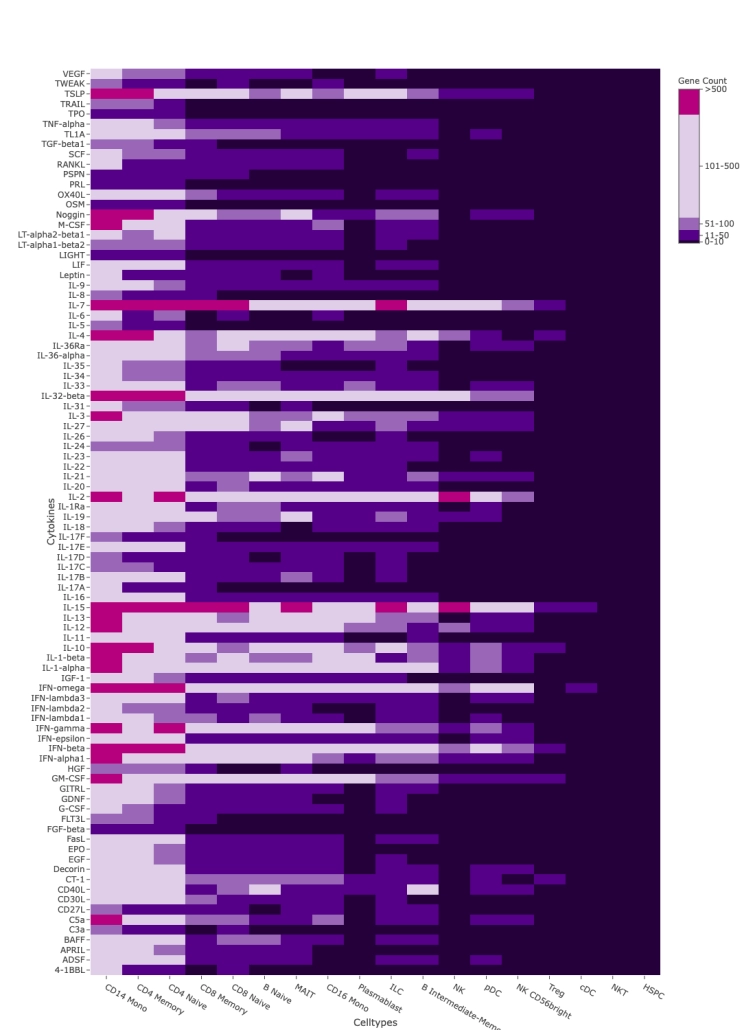
Figure 3: Cytokine-Induced Gene Changes
A heatmap summarizes the averaged number of genes significantly upregulated (log fold change >0.3, p <0.001) for each cell type and cytokine, highlighting which immune cells respond most strongly to specific cytokine treatments.
Parse 10M PBMC Cytokines Clustering Tutorial
Joey Pangallo, Efi Papalexi – Parse Biosciences, Seattle, WA
Step-by-step example of analyzing 10 million PBMCs treated with cytokines using the Evercode workflow. Covers data loading, preprocessing, Leiden clustering, and generating UMAP plots with Scanpy.
Parse 10M PBMC Cytokines Clustering Tutorial (Downsampled)
Joey Pangallo, Efi Papalexi – Parse Biosciences, Seattle, WA
Same workflow as above, starting with a downsampled dataset of 1 million cells. Ideal for quicker exploration or limited CPU memory setups.
scCODA Parse 10M PBMC Cytokines
Artur Szałata, Dominik Klein, Soeren Becker, Fabian Theis – Helmholtz-Munich
Demonstrates analysis of cell proportion changes across 10 million PBMCs. Shows how using the full dataset improves statistical significance of perturbation effects. Based on scCODA, a Bayesian model for compositional single-cell data analysis (Nat Commun 12, 6876, 2021).
Parse 10M PBMC Cytokines Dask Workflow
Artur Szałata, Dominik Klein, Soeren Becker, Fabian Theis – Helmholtz-Munich
Walks through preprocessing the 10M cell dataset using Dask. Loads data chunk-wise to reduce memory use and demonstrates highly variable gene selection for downstream analysis.
Dataset License: CC BY-NC 4.0 (non-commercial use). Commercial licensing inquiries: support@parsebiosciences.com

ANZ Market Manager - Research Genomics
This site uses cookies. By continuing to browse the site, you are agreeing to our use of cookies.
OKLearn moreWe may request cookies to be set on your device. We use cookies to let us know when you visit our websites, how you interact with us, to enrich your user experience, and to customize your relationship with our website.
Click on the different category headings to find out more. You can also change some of your preferences. Note that blocking some types of cookies may impact your experience on our websites and the services we are able to offer.
These cookies are strictly necessary to provide you with services available through our website and to use some of its features.
Because these cookies are strictly necessary to deliver the website, refusing them will have impact how our site functions. You always can block or delete cookies by changing your browser settings and force blocking all cookies on this website. But this will always prompt you to accept/refuse cookies when revisiting our site.
We fully respect if you want to refuse cookies but to avoid asking you again and again kindly allow us to store a cookie for that. You are free to opt out any time or opt in for other cookies to get a better experience. If you refuse cookies we will remove all set cookies in our domain.
We provide you with a list of stored cookies on your computer in our domain so you can check what we stored. Due to security reasons we are not able to show or modify cookies from other domains. You can check these in your browser security settings.
These cookies collect information that is used either in aggregate form to help us understand how our website is being used or how effective our marketing campaigns are, or to help us customize our website and application for you in order to enhance your experience.
If you do not want that we track your visit to our site you can disable tracking in your browser here:
We also use different external services like Google Webfonts, Google Maps, and external Video providers. Since these providers may collect personal data like your IP address we allow you to block them here. Please be aware that this might heavily reduce the functionality and appearance of our site. Changes will take effect once you reload the page.
Google Webfont Settings:
Google Map Settings:
Google reCaptcha Settings:
Vimeo and Youtube video embeds:
The following cookies are also needed - You can choose if you want to allow them:
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.