Our STOmics validated tissue list provides researchers with a comprehensive reference of hundreds of tissue types successfully tested using Stereo-seq, the cutting-edge spatial transcriptomics technology. Each tissue entry includes detailed sample information, experimental parameters, and test results, allowing scientists to make informed decisions before starting their single-cell or spatial transcriptomics experiments. By consulting this list, you can ensure compatibility with your tissue samples and streamline your experimental design.
The list not only highlights tissue types that have been validated but also provides insights into the experimental conditions that yielded the most reliable results. Researchers can leverage this information to optimize sample preparation, sequencing protocols, and data quality control measures. This reduces trial-and-error, saves valuable time and resources, and ensures reproducibility across studies. It is an essential tool for anyone planning to use Stereo-seq for spatial gene expression profiling.
In addition, our STOmics validated tissue list supports better planning for large-scale studies and comparative analyses. By providing a centralized reference for tissue performance, it helps guide tissue selection, anticipate potential challenges, and maximize experimental success. Whether you are exploring new tissue types or scaling up existing workflows, this validated tissue list is your key resource for robust, high-quality spatial transcriptomics research.
Notices:
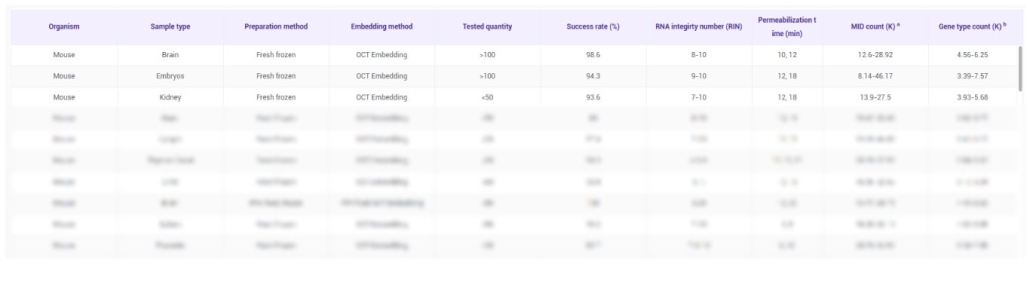
The STOmics validated tissue list was generated using standard tissue and sample types, all of which are frozen. Each tissue sample had an area of less than 1 cm² and was sectioned at a thickness of 10 μm. Most experiments were performed using the Stereo-seq Transcriptomics Kit V1.2, with a few using V1.1. Sequencing depth ranged from 1–3 G reads per sample, and data were processed using the Stereo-seq Analysis Workflow (SAW) versions V2.1.0–V5.1.3. Testing was conducted between 2020 and 2022.
Please note that all test parameters and results are highly dependent on the tissue and sample type. This information should be used as a reference guide to help design and optimize your own experiments, rather than as definitive outcomes for all samples.
Key parameters included in the list:
MID (K): Bin200_Median_MID in thousands
GENETYPE (K): Bin200_Median_Genetype in thousands

ANZ Market Manager - Research Genomics
This site uses cookies. By continuing to browse the site, you are agreeing to our use of cookies.
OKLearn moreWe may request cookies to be set on your device. We use cookies to let us know when you visit our websites, how you interact with us, to enrich your user experience, and to customize your relationship with our website.
Click on the different category headings to find out more. You can also change some of your preferences. Note that blocking some types of cookies may impact your experience on our websites and the services we are able to offer.
These cookies are strictly necessary to provide you with services available through our website and to use some of its features.
Because these cookies are strictly necessary to deliver the website, refusing them will have impact how our site functions. You always can block or delete cookies by changing your browser settings and force blocking all cookies on this website. But this will always prompt you to accept/refuse cookies when revisiting our site.
We fully respect if you want to refuse cookies but to avoid asking you again and again kindly allow us to store a cookie for that. You are free to opt out any time or opt in for other cookies to get a better experience. If you refuse cookies we will remove all set cookies in our domain.
We provide you with a list of stored cookies on your computer in our domain so you can check what we stored. Due to security reasons we are not able to show or modify cookies from other domains. You can check these in your browser security settings.
These cookies collect information that is used either in aggregate form to help us understand how our website is being used or how effective our marketing campaigns are, or to help us customize our website and application for you in order to enhance your experience.
If you do not want that we track your visit to our site you can disable tracking in your browser here:
We also use different external services like Google Webfonts, Google Maps, and external Video providers. Since these providers may collect personal data like your IP address we allow you to block them here. Please be aware that this might heavily reduce the functionality and appearance of our site. Changes will take effect once you reload the page.
Google Webfont Settings:
Google Map Settings:
Google reCaptcha Settings:
Vimeo and Youtube video embeds:
The following cookies are also needed - You can choose if you want to allow them:
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.