PRODUCTS
The Twist NGS Methylation Detection System provides a robust, end-to-end sample preparation solution for identifying methylated regions in the human genome. The workflow employs a unique enzymatic process from New England Biolabs® that is much less damaging to DNA, alongside Twist‘s Custom Methylation Panel design.
Whether you are investigating cellular differentiation or screening liquid biopsies for cancer, the system offers the most efficient methylation detection available.
The workflow features enzymatic conversion of unmethylated cytosines (see figure) to identify sites of methyl-cytosine (5mC) and hydroxymethyl-cytosine (5hmC).
Enzymatic conversion produces more intact libraries with better representation, and ultimately achieves more sensitive methylation detection. The library preparation system is suitable for whole genome sequencing and downstream enrichment with Twist Methylation Panels.
EM-seq conversion involves a series of enzymatic steps to convert unmethylated cytosines into uracils. The final result is the same as conventional bisulfite conversion, making EM-seq compatible with existing analysis pipelines that use Bismark and bwa-meth.
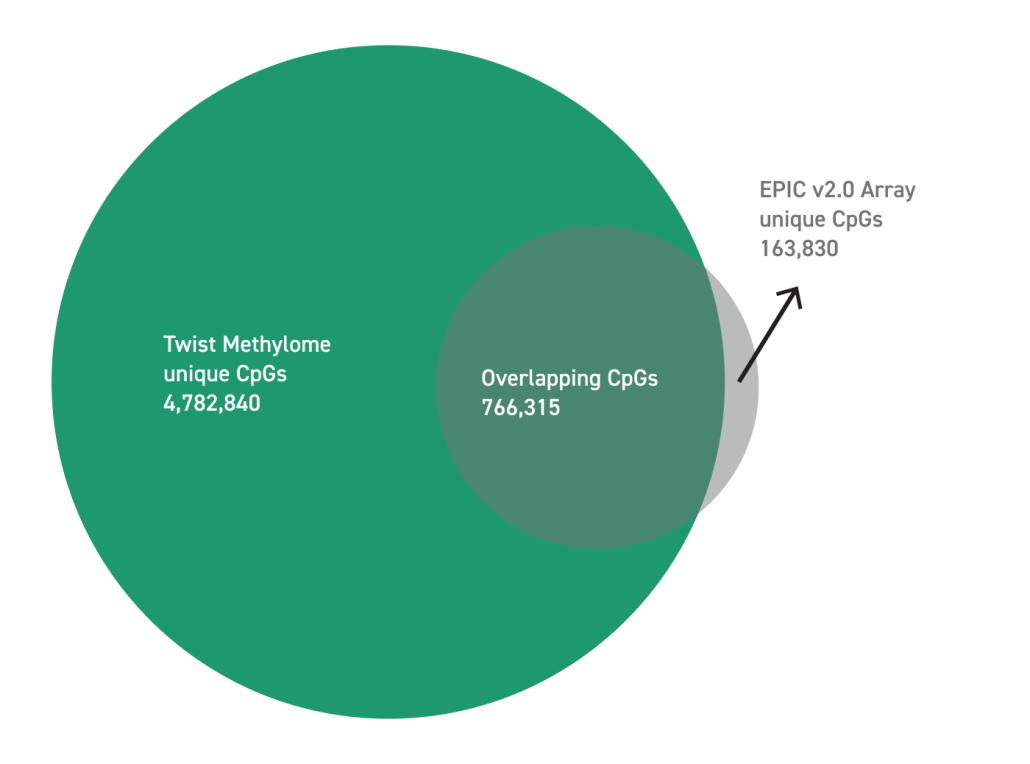
The Twist Human Methylome Panel targets 3.98M CpG sites through 123 Mb of genomic content to target biologically relevant methylation markers. Expansive content makes this panel an ideal choice for investigators to explore the methylation fraction in a diverse range of applications from cancer metastasis, human development, and functional genomics.
The panel is optimized and validated for use with the Twist methylation detection system for a complete end-to-end workflow with industry leading performance. High capture efficiency increases the sensitivity of detection across the footprint of the epigenome while decreasing sequencing costs. The panel is ideal for screening cohort samples and differentially methylated region discovery.
The Twist Human Methylome Panel offers comprehensive coverage of the genome, targeting 3.98 million CpG sites within 123 Mb of genomic content. It efficiently identifies 84% of CpG islands and covers an additional 105,288,339 bases of related regions. In comparison to average microarrays, the panel overcomes static content limitations and enhances coverage across the epigenome. Microarrays suffer from constraints in methylation detection at extreme ends due to background noise and saturation, whereas the Methylome Panel, utilizing hybrid-capture panels and Next-Generation Sequencing (NGS), provides expanded content, single-base resolution, and a higher dynamic range for more accurate detection of differentially methylated regions. The panel’s performance is notable, achieving 90% coverage at 30x depth, 95% on-target rates, and high uniformity with a fold 80 of 1.54. Its efficient capture metrics instill confidence in accurate methylation fraction detection while minimizing sequencing costs.
Get a call from your local Decode Science representative to help you find the best fit genomics products for you.
Or give us a call at:
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.
Simply add your details below and get quick access to the brochure.